Systems Medicine
of Infectious Diseases
Research and Consulting Group
Machine Learning for Multiscale Analysis of Biomedical Data
A combined modeling framework to break the lethal alliance between influenza and bacterial coinfections
The 1918 influenza pandemic derived about 50 million deaths (approximately 3-5% of the world's population). Retrospective studies performed on victims of the devastating 1918/1919 influenza pandemic and the recent H1N1 pandemic revealed a high incidence of coinfections with unrelated bacterial pathogens. In fact, 70% of the alarmingly high death toll during the 1918/1919 outbreak was attributed to coinfection with Streptococcus pneumoniae. Several mechanisms have been implicated in the viral-bacterial synergism, which altogether demonstrates a multifactorial and complex nature of co-pathogenesis. However, it is still largely unknown what the relative contribution of the innate immune response and consequently to bacterial outgrowth in secondary infection is and whether immune systems components work alone or in synergism as `friend or foe' to the coinfected host.
In this project, by combining tailored in vivo mouse infection experiments at the Otto von Guericke University Magdeburgg and computational methods established at the Frankfurt Institute for Advanced Studies, we aim at clarifying the role of Natural Killers and CD8+ T cells, and the relative contribution of inflammatory cytokines to impair macrophages functions on enhanced susceptibility to bacterial coinfection following influenza.
Sponsor: DFG
Period: 2018 - 2022
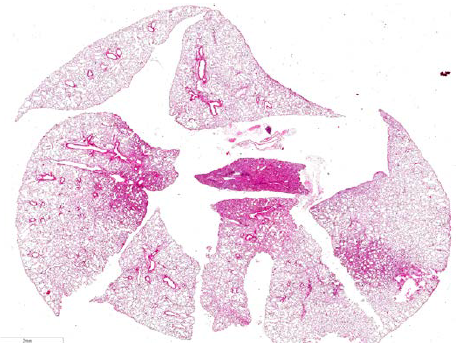
Figure: Ηistopathological changes in the lungs of mice infected with influenza virus